Artificial Intelligence
Process High Resolution Tissue images in Medical Domain for Model Training
The digitization of histopathological slides through Whole Slide Imaging (WSI) is revolutionizing pathology by enabling ultra-high-resolution digital scans for precise disease diagnosis and treatment planning. While WSI facilitates AI-driven analysis and remote diagnostics, its adoption faces challenges due to massive image dimensions and computational constraints. Conventional down-sampling approaches often compromise critical diagnostic details, reducing model accuracy. This paper proposes an intelligent preprocessing strategy that segments WSIs into manageable, high-information tissue patches, preserving essential features for deep learning models. By optimizing patch selection and reducing redundancy, the method enhances diagnostic accuracy and scalability, paving the way for robust AI-assisted pathology in resource-limited and high-throughput healthcare environments.
Insights
- Whole Slide Imaging (WSI) is transforming pathology by digitizing entire microscope slides into gigapixel-scale images, enabling detailed tissue analysis for AI-driven diagnostics.
- WSIs introduce major technical challenges: massive file sizes (up to 100,000×100,000 pixels), high storage demands, and complex tissue morphology that exceeds conventional GPU and memory limits.
- Naive resizing or dense tiling approaches either lose critical diagnostic details or create computational bottlenecks, making traditional deep learning pipelines ineffective.
- This white paper explores an intelligent preprocessing method that extracts high-information tissue patches, reducing data volume while preserving diagnostic features for accurate AI models.
- Readers will gain insights into best practices for WSI patch extraction, computational trade-offs, and applications in healthcare (e.g., cancer diagnostics, rare disease detection) and manufacturing (e.g., metal surface defect detection).
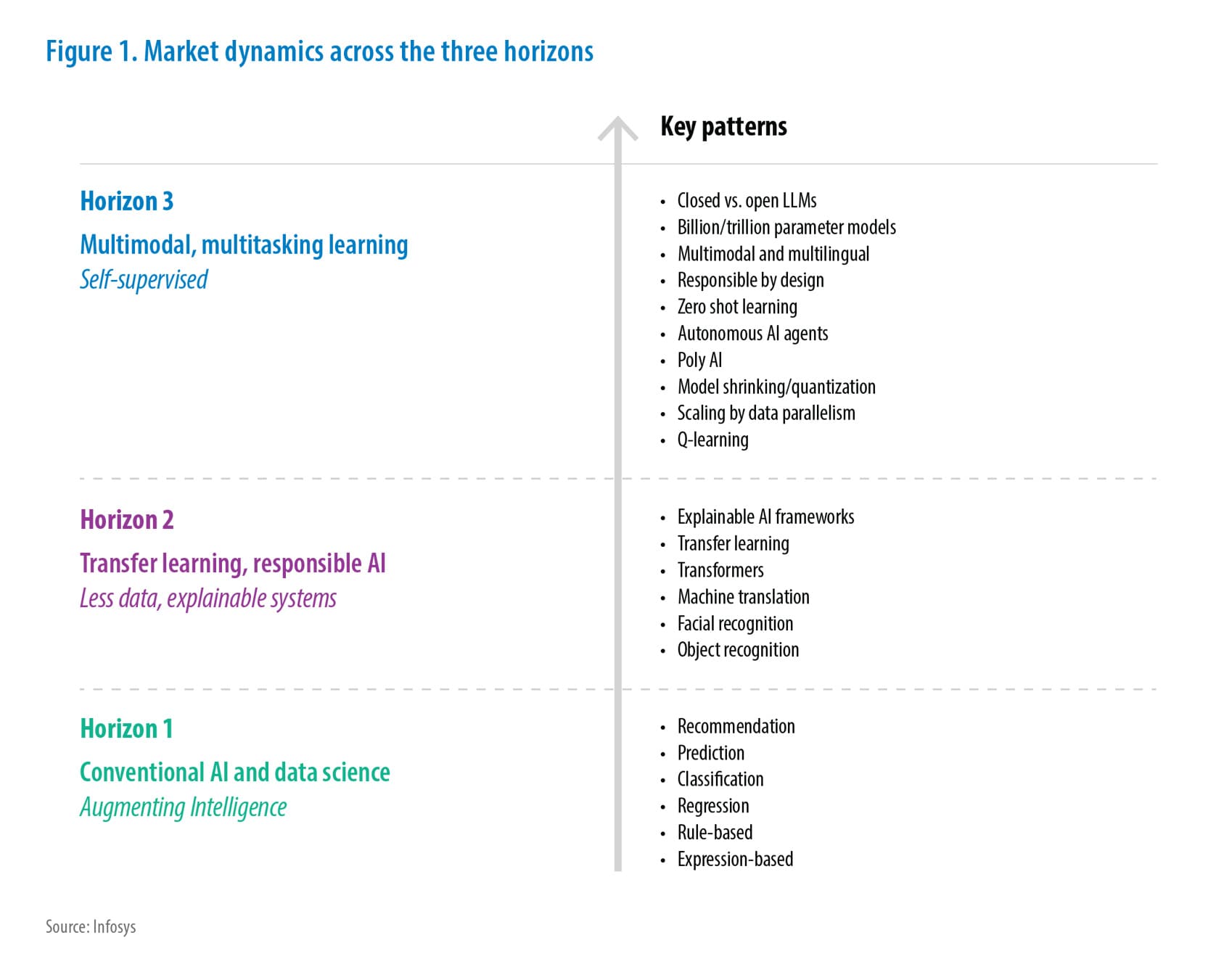
Whole-slide imaging (WSI) has revolutionized pathology by digitizing entire microscope slides for analysis. Figure 1 shows an example: a gigapixel-scale histology slide. Such slides can span tens of thousands of pixels per side (e.g. ~100,000×100,000 at high magnification pmc.ncbi.nlm.nih.gov) and occupy multiple gigabytes (20× scans ≈0.8 GB, 40× ≈1.2 GB en.wikipedia.org). This is roughly 100–1000× more data per image than typical radiology scans en.wikipedia.org. These ultra-high-resolution WSIs enable detailed tissue examination but pose severe technical challenges. Naively resizing WSI’s for deep learning leads to loss of important diagnostic detail. At the same time, AI in pathology is gaining traction – recent studies show deep learning models can achieve pathologist-level performance on slide interpretation nature.com. To bring this promise into routine digital healthcare, we must overcome the data size hurdles (talked below) making sure we are also capturing enough features for any deep learning model to learn with high accuracy which is clinical demand.
Fig 1: Example whole-slide image (WSI) of a pathology slide (hematoxylin & eosin stain)

- Gigapixel scale: Whole-slide scans typically contain billions of pixels. For example, Figure 1’s slide is roughly an entire tissue section at high magnification. Processing such data directly far exceeds typical GPU or memory capabilities.
- Massive storage: Digital pathology requires on the order of 100–1000× the storage of conventional radiology. A single patient’s slides can quickly accumulate hundreds of gigabytes.
- Complex morphology: WSIs show extreme tissue heterogeneity and common artifacts (staining variations, folds, out-of-focus regions). This “what and where” problem – identifying subtle patterns in a massive image – impedes straightforward CNN analysis.
- Clinical demand: Pathologists need fast, accurate tools. Automated analysis of WSIs can dramatically reduce workload and turnaround times, but only if algorithms are both accurate and efficient.
In short, while AI-driven analysis holds great promise in digital pathology, conventional deep learning pipelines break down on whole-slide data. Down sampling and resizing a WSI to feed a CNN, for example, may retain global appearance but blurs or eliminates cellular details needed for diagnosis. Conversely, naively tiling a slide into dense patches explodes the number of images and computations. Therefore, we must intelligently reduce WSI data volume without sacrificing tissue-level information with preprocessing method, which explained in the paper. By examining this technique, readers gain insights into best practices for WSI patch extraction, understand the computational trade-offs involved, and explore potential applications in health care areas like cancer diagnostics, rare disease detection, and AI-assisted pathology research. Owing to its general applicability Further its application can also be extended to Manufacturing use cases like Identification of Metal Surface defects which might require careful analysis of metal sheets for hairline cracks, narrow dents which can lead to defective finished products down the assembly & finishing line.
Introduction
In the evolving landscape of digital healthcare, the digitization of histopathological slides through Whole Slide Imaging (WSI) has emerged as a pivotal advancement. It is transforming the field of pathology by enabling clinicians and researchers to engage with tissue samples in a more interactive, scalable, and precise manner. WSI provides ultra-high-resolution digital scans of entire pathology slides — often reaching dimensions like 5000×20000 or higher — that preserve microscopic details critical for detecting cellular anomalies, diagnosing diseases, and formulating treatment strategies.
As healthcare systems push toward more data-driven and AI-assisted diagnostic models, WSI opens new possibilities for automation, reproducibility, and remote diagnostics. The ability to process and analyze these digital slides using advanced computer vision and deep learning algorithms offers not only a step forward in diagnostic accuracy but also the potential to scale expert-level analysis across under-served regions and high-throughput medical settings.
However, this promise remains only partially realized.
Despite rapid progress in artificial intelligence, the application of deep learning to WSIs presents significant technical and infrastructural challenges. These challenges stem primarily from the extremely high resolution and large file sizes of WSI data, which introduce computational bottlenecks during model training and inference. Conventional approaches like simple resize down sampled (Explained Latter) WSI into manageable size like 1024/1024 or 1536/1536 does mitigate the problem of handling high resolution and sizing but it often sacrifice critical diagnostic information , leading to suboptimal accuracy by any Deep Learning model trained with those ,same is demonstrated by Experiment down in the paper. Additionally, many regions within a WSI may contain redundant or non-informative or empty background areas, making it imperative to focus only on diagnostically relevant tissue structures.
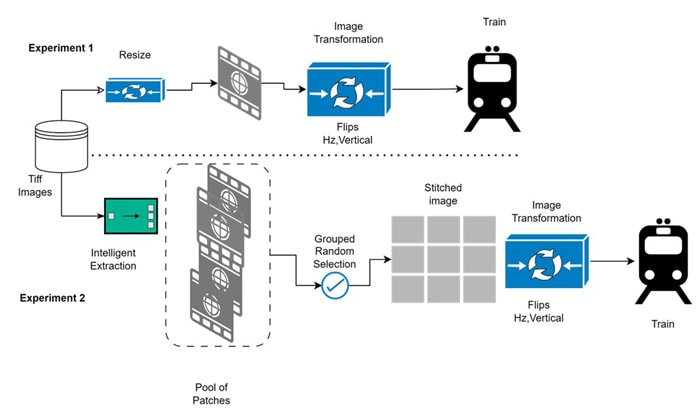
This White paper talks addressing these challenges through a preprocessing approach that not only breaks down large size WSI into smaller sizes that can be easily processed with available Compute but also intelligently extract tissue patches with detailed features through which Computer vision model can learn definite tissue feature pattern that can form basis to detect the ailment .By efficiently selecting meaningful patches of manageable size, this method enhances the feasibility of using WSIs for AI-driven medical diagnosis, ultimately contributing to more accurate and scalable pathology applications , even in the face of massive image dimensions and hardware constraints.
Proposed Preprocessing Method
The preprocessing pipeline consists of the following key steps (pseudocode below). It takes a whole-slide image as input and outputs a list of high-quality tissue patch coordinates which can then be used to extract the patch images to be used for training of Deep learning model:
Reading Image
Generally, the WSI images are in the TIFF format they can read through 2 commonly used Python libraries.
- Tiff images can be read using two prominent libraries which allows us to read desired regions as per provided X,Y locations
- skimage.io.MultiImage: Offers image reading with different sizes referred as level of dimension with respect to original Dimensions of image . Sizes vary upon extent of down sampling we chose like original,4th or 8th meaning ¼ or 1/8th of original image size.
- openSlide.openSlide : Same as skimage enables the reading of tiff file, difference being to read region one needs to provide X,Y location with respect to Original Image size compared to down sampled image size in case skimage library.
Fig 2: Shows Whole slide image of Tissue obtained through Biopsy for Diagnosis of concerned ailment.

Understanding Down Sample
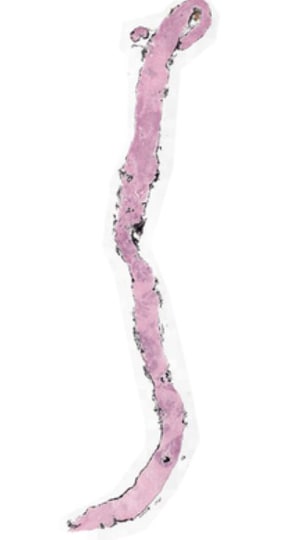
Understanding Down sampling: Down Sampling is a process to take the high-resolution Whole Slide Image (WSI) and get lower lever representations of the slide. Higher the down sampling rate less resolution the image will have (needs less memory and compute) . As shown above in the Meta data of Tiff files Fig2 there are three levels of dimensions each dimension is result of down sample factor 1 meaning original, 4 meaning 1/4th of original and 1/16th meaning 1/16 of original. Often Down sampling becomes necessary when you have limited computing at hands and you may want to cover more features area for a given patch size ,offering better contextuality without significant degradation of image quality.
We can also see in the fig 3 below height/width of image start getting fractioned depending upon down sample factor of dimension level that one chose so I memory footprint getting reduced that would mean less computer resourcing needs.
Fig 3: WSI of the Biopsy tissue
Original Image can be down sampled to factor depending upon Compute resources at hand. Based on experiments done for checking feature granularity needs (lesser the patch size more granularity but more number of patches to process) and amount of computing at hand on an average & number of patches that could sufficiently meet feature coverage needs for a given patch size Preferable down sample factor is 1/4th .This is again a hyper parameter that can be adjusted for a given use case depending upon images, features coverage required. Typical Patch size that can work are 256-512 with down sampling rate of 1/4th.
Tissue Masking
The WSI is first down-sampled (Explained above) and color-thresholded (or processed via deep models) to produce a binary mask of tissue vs. background. For example, grayscale thresholding on a low-resolution copy can efficiently separate stained tissue (foreground) from glass (background) across the slide.
Mask Refinement
We apply morphological dilation and contour smoothing to close holes and smooth edges in the raw mask. This ensures contiguous tissue blobs and avoids spurious gaps.
Fig 4: WSI Slide and Corresponding Refined Mask

Padding & Segmentation
The refined mask is padded at its borders so that tissue at the very edge is not lost. Then we identify connected components (contours) in the mask – each corresponds to a contiguous tissue region (e.g. one tissue fragment or section).
Smart Patching
Rather than a uniform grid, we sample fixed-size patches within each tissue segment. Patches are placed to cover the segment efficiently (e.g. using a sliding window or quadtree on that region), focusing only on tissue areas. This yields a set of high-resolution patches that span the entire tissue content without needless overlapping or empty background.
Patch Evaluation
Each candidate patch is scored by its tissue content (for example, the fraction of non-background pixels) and other quality metrics. Patches failing a threshold of tissue density or clarity are discarded.
Final Output
The pipeline returns the coordinates of the remaining high-quality patches. These patches retain the full detail of the original slide in tissue regions, ready for AI analysis. These coordinates can then be used to extract the patches from original WSI slide through openSlide or skimage. Extracted patch through selected coordinates would look like as shown in the fig5 below, while fig 4 shows intelligent selection of patches leaving out the unnecessary background white information that may occupy at least 75 percent of whole image.
Fig 5: Applying the Intelligently selected coordinates on original slide covering only tissue content
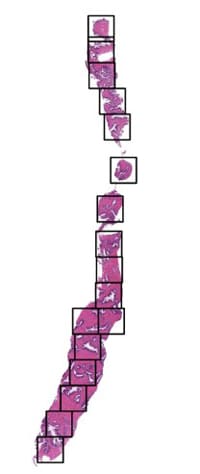
Fig 6: Extracted Patches from Original Slide
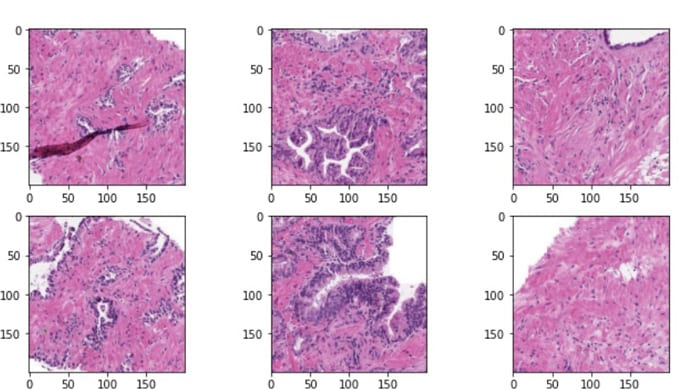
Using this method, we pass intelligently extracted some N Stitched patches each with size 512×512–1024×1024 patches per slide (for example) in every iteration of model training. Critical tissue features – like deep inner gland structures, potentially inflammatory cells features etc. – remain fully preserved. By comparison, a simple resize-to-512-1024 approach would smear out these details, severely degrading diagnostic performance.
Experiment
We validated the proposed method on a large-scale pathology dataset (~418 GB of WSI data) associated with Prostate gland-stained biopsy slides for cancer grade detection, comparing it against a baseline “resize-only” approach. In both experiments we used standard CNN architectures and measured performance with Cohen’s Quadratic Weighted Kappa (QWK), a common metric for ordinal classification/agreement in pathology.
Experiment 1 – A multiclassification experiment where in objective was to develop deep learning models for detecting Prostate Cancer (PCa) on images of prostate tissue samples and estimate severity grade of the disease between 0-5.
Given was Dataset with High Resolution Tissue image size 418GB
- Pytorch Dataset was prepared to read Tiff images using skimage and process Tiff image further by simply resizing the image to 256/256 size using open CV resize method.
- Resized images were used to Train Se-Resnext50 model.
- Evaluation metric Quadratic cohen_kappa_score from sklearn.metric was used to validate the model.
- Validation set QWK Score obtained was 64.34 and Test data QWK score obtained was 62.95.
Experiment 2 – Same Experiment was performed using Resnext50 model as deep learning model and patching approach above as data preprocessing method.
- Given was Dataset with High Resolution Tissue image size 418GB.
- Using the above approach Every single Tiff image was divided into the multiple patches of size 256.
- Pytorch Dataset was prepared to sequentially select from randomly selected formed Groups (like continuous selection from first N patches, Middle N patches, Ending N Patches) the prepared patches (using proposed preprocessing method) and those stitched together to make single image of desired size like 1536.
- Stitched images were used to Train Resnext50 model . Every iteration different group was selected during the Training to cover the whole slide for diagnosis.
- Evaluation metric Quadratic cohen_kappa_score from sklearn.metric was used to validate the model.
- Validation set QWK Score obtained was 88 and the Test data QWK score obtained was 92.
Additional details on Training:
- Training Data & Problem statement: https://www.kaggle.com/competitions/prostate-cancer-grade-assessment/data
- Augmentation: Albumentation’s Normalize, HorizontalFlip, VerticalFlip
- Train/Test Split: Stratified Kfold -4 folds on Training Target column ISUP_GRADE
- Batch size: 16-24 (Depending upon GPU memory)
- Epochs: 10-15
- Learning rate: 1e-3
Fig 7: Training Pipeline flow
Comparison
These results demonstrate the advantage of preserving tissue detail: the patch (extracted through the preprocessing method) based model’s predictions aligned much more closely with ground truth (QWK ≈88–92%) than the resize-only model. Note that QWK values close to 1.0 indicate nearly perfect agreement, so the gains here are meaningful for diagnostic accuracy. (For context, prior pathology AI studies report QWK in a similar range when expert-level performance is reached. Thus, we can clearly find that explained Data Preprocessing leads to much better Accuracy compared to the base line approach which uses just normal resizing of the down-sampled original image with same factor in both cases. Analysis of such images for diagnosing the ailments in field of medical domain needs deep understanding of Tissues features which are done today manually under the microscope with enlargement of area to be in focus. Intelligently selected tissue Patching Tries to simulate similar things by providing enlarged view of various sections of Tissue and hence results in better Model learning.
Impact & Benefits
This efficient preprocessing has profound practical implications:
- Scalability: By discarding blank regions, the pipeline cuts the number of input pixels by orders of magnitude. A WSI that might have required hundreds of GBs can be represented by a few hundred patches (each only megapixels). This makes it feasible to process and store data from large slide collections, and to deploy high-resolution AI on standard GPUs.
- Speed and Resource Efficiency: Fewer total pixels means much faster inference. Models run on dozens of 512×512 tiles instead of a multi-gigapixel image, dramatically reducing memory and compute requirements. This enables high-throughput pipelines (e.g. analyzing hundreds of slides per day) without supercomputing resources.
- Accuracy and Reliability: Retaining tissue detail avoids the smoothing artifacts of resizing. Our experiments show that diagnostic agreement (QWK) improves substantially, meaning AI predictions are more reliable. In practice this could translate to fewer missed findings and more consistent grading.
- Clinical Benefits: Faster, more accurate AI support can speed diagnosis and treatment. As noted in the literature, digitized slide analysis has the potential to “decrease the workload of pathologists, reduce turnaround times for reporting, and standardize clinical practices. Our pipeline directly contributes to these goals by making automated analysis practical on a scale.
- Accessibility: Smaller patch data means slides or features can be transmitted more easily (for telepathology or multi-center studies) and processed in resource-constrained settings (e.g. community hospitals or remote labs). This broadens the reach of advanced pathology AI.
- Resource Utilization: By leveraging existing deep networks (e.g. ResNeXt50) on intelligently preprocessed data, institutions can use off-the-shelf hardware and models. There is no need for exotic memory systems or bespoke scanners – the innovation is entirely in smarter software.
Briefs on Advances in Area
As recent advances made into patch extraction techniques from WSIs, work by X et al., 2025 introduced an Abnormality-Aware Multimodal Learning (AAMM) framework that enhances classification by integrating visual and textual modalities. The method employs a Gaussian Mixture Variational Autoencoder (GMVAE) to prioritize abnormal regions during patch selection, followed by a cross-attention mechanism that fuses patch-level, cell-level, and text-derived features from LLAVA (VLM). This approach has demonstrated improved performance in tumor classification and subtyping tasks and can be considered a complementary screening layer post initial patch extraction to refine diagnostic focus by further reducing number of images to use for model training or inferencing.
Extensibility to other Domains
While the proposed preprocessing pipeline is explained for pathology, its core principles are broadly applicable to any domain dealing with ultra-high-resolution images that require selective and intelligent analysis. Many industries beyond healthcare – particularly those in manufacturing, materials science, defense, and satellite imaging – face similar challenges: large image sizes, small regions of interest, and the need to extract meaningful insights from fine-grained patterns within a massive visual space. Thus, one can bring in the automated visual inspection for various manufacturing use cases such as detecting microscopic defects in metal sheets, composite materials, semiconductors, or printed circuit boards (PCBs). Below Table provides the potential application of the explained preprocessing method for use cases across various domains.
| Domain | Analogous Use Case | Benefit of Smart Preprocessing |
|---|---|---|
| Aerospace Manufacturing | Surface flaw detection in turbine blades or composites | Focus analysis on material defects, cracks, or delamination zones |
| Electronics/PCB QA | Detecting soldering issues or misalignments | Targets high-density component zones and reduces false positives |
| Materials Science | Microstructure analysis (e.g. grain boundaries, phase distribution in alloys) | Maintains detail in micro-regions for texture-based modeling |
| Satellite Imagery | Urban planning, crop health, or military reconnaissance | Segments out clouds, oceans, or forests to focus on man-made zones |
| Archaeology/ Forensics | Digitized artifact examination or forensic trace evidence | Isolates regions with engravings, micro-fibers, or trace materials |
Conclusion
Overall, the paper explores efficient patch extraction techniques that can help Data Scientists train deep learning models through WSIs having high resolutions effectively without being constrained by hardware limitations. This approach identifies and extracts diagnostically relevant regions, ensuring that critical pathological features are retained. This targeted method improves both computational efficiency and model accuracy as Model get visibility to those deep features which are examined under the Microscope manually, making AI-driven pathology more scalable & precise. By learning this technique, readers gain insights into the best practices for WSI patch extraction followed by better Model training, understand the computational trade-offs involved, and explore potential applications in cancer diagnostics, rare disease detection, and AI-assisted pathology research.
References
Throughout the preparation of this whitepaper, information and insights were drawn from a range of reputable sources, including research papers, articles, and resources. Some of the key references that informed the content of this whitepaper include:
- https://pmc.ncbi.nlm.nih.gov/articles/PMC6882930/#:~:text=methodologies%20on%20visual%20understanding,In%20this%20paper%2C%20we
- https://pmc.ncbi.nlm.nih.gov/articles/PMC6882930/#:~:text=pathologies.%20However%2C%20WSIs%20are%20multi,by%20the%20morphological%20variance%2C%20artifacts
- https://en.wikipedia.org/wiki/Digital_pathology#:~:text=Besides%20this%20difference%20in%20pre,similar%20to%20those%20in%20radiology
- https://pmc.ncbi.nlm.nih.gov/articles/PMC6882930/#:~:text=Recently%2C%20NHS%20Greater%20Glasgow%20and,Furthermore%2C%20deep%20learning%20techniques
- https://www.researchgate.net/figure/Left-to-Right-An-illustration-of-the-tissue-mask-overlayed-on-a-small-region-of-the-WSI_fig2_352065999#:~:text=...%20pipeline.%20The%20pre,
- https://pmc.ncbi.nlm.nih.gov/articles/PMC6882930/#:~:text=Recently%2C%20NHS%20Greater%20Glasgow%20and,Furthermore%2C%20deep%20learning%20techniques
- https://www.kaggle.com/competitions/prostate-cancer-grade-assessment
- https://www.kaggle.com/code/akensert/panda-optimized-tiling-tf-data-dataset

Subscribe
To keep yourself updated on the latest technology and industry trends subscribe to the Infosys Knowledge Institute's publications
Count me in!